Reverse transcription reaction, as one of the basic techniques in molecular biology research, has been widely used in several research fields and can be regarded as the watchword for researchers to conduct RNA function studies.
There are six known areas of application for reverse transcription:
- Reverse Transcription Polymerase Chain Reaction (RT-PCR)
- Quantitative RT-PCR (RT-qPCR)
- cDNA cloning and library construction
- Rapid Amplification of cDNA ends (RACE)
- Gene expression microarrays
- RNA Sequencing (RNA-Seq)
- Reverse transcription polymerase chain reaction (RT-PCR)
In RT-PCR, reverse transcription is one of the key steps in the success of the experiment as reverse transcription converts RNA to cDNA, which is then amplified by the PCR reaction to provide a template for downstream experiments (figure 1). The selected reverse transcription should have the highest efficiency for all samples, even difficult-to-transcribe RNA samples such as those that are degraded, have inhibitor residues, or have a high degree of secondary structure.
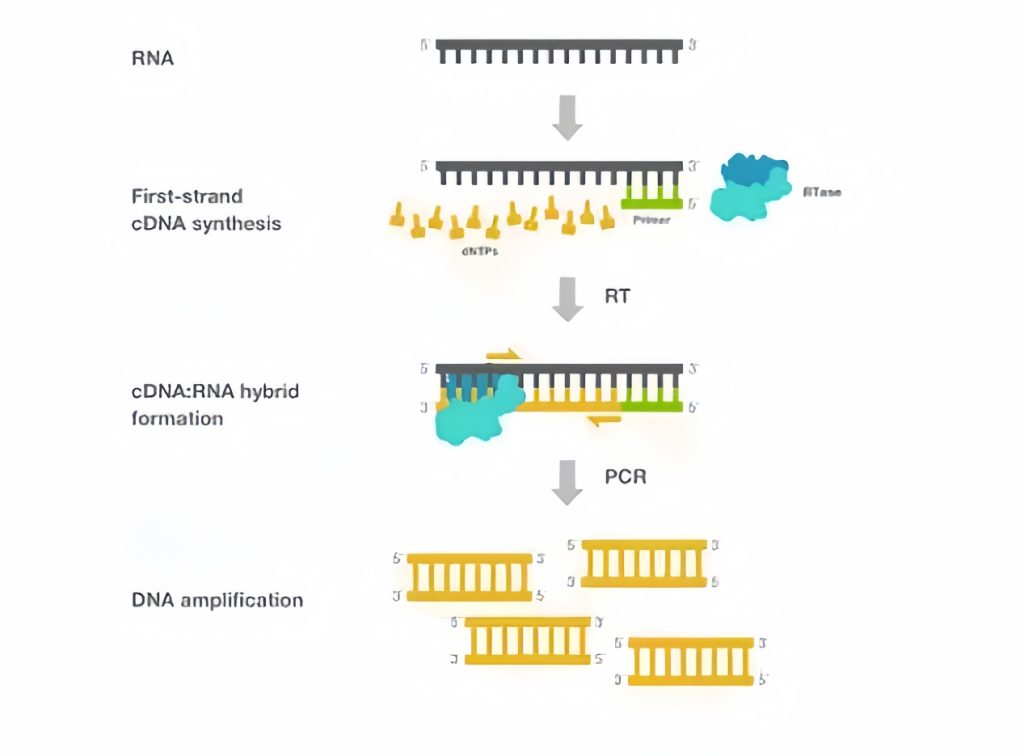
Figure1. Reverse transcription polymerase chain reaction (RT-PCR). rt = reverse transcription, rtase = reverse transcription
- Quantitative RT-PCR (RT-qPCR)
One of the most common applications of quantitative RT-PCR (RT-qPCR) is through the quantification of real-time mRNA levels in cells and tissues over a period of time or after an event (e.g., drug treatment). RT-qPCR is more sensitive than RT-PCR, and the accuracy of RT-qPCR gene expression quantification depends largely on the quality and quantity of the cDNA template. Therefore, reverse transcription is critical to the success of RT-qPCR. The selected reverse transcription should have the following characteristics:
- Ability to synthesize cDNA efficiently, even for low abundance genes as well as sub-quality and/or difficult-to-transcribe RNA samples.
- Optimal dynamic range or linearity of cDNA over a wide range of RNA starting amounts ensures accurate gene expression quantification.
- The selected reagents should produce high and consistent cDNA yields during amplification to obtain gene expression results with high sensitivity and low variability. Reverse transcription in premixed form can be considered to minimize experimental errors.
A special procedure for RT-qPCR is direct reverse transcription from crude cell lysates without RNA isolation. In experiments where scarce samples are used or where specific cells within a population are selected, the direct RT-qPCR method may be considered to prevent possible sample loss and low RNA recovery.
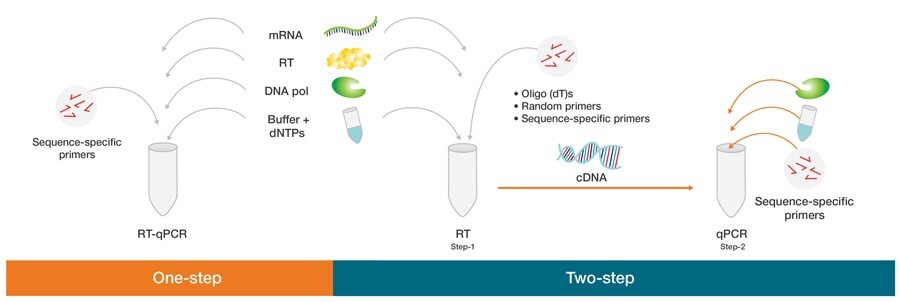
Figure2. One-step vs. two-step RT-qPCR.
- cDNA cloning and library construction
A cDNA library may consist of cDNA clones representing transcribed sequences from a specific sample. Thus, the library provides spatiotemporal gene expression information about a particular cell type, organ, or developmental stage. cDNA library clones can be used to identify novel RNA transcripts, determine gene sequences, and recombinant protein expression.
A necessary condition for the construction of cDNA libraries is that the RNA is appropriately representative of its full length and/or relative abundance, and therefore, the choice of reverse transcriptase is important. Reverse transcriptase with high sustained synthetic capacity can synthesize long cDNAs and capture low abundance RNAs. similarly, when reverse transcribing RNAs with a high degree of secondary structure, it is recommended to use reverse transcriptase with high thermal stability
- Rapid amplification of cDNA ends (RACE)
Rapid amplification of cDNA ends (RACE) is a PCR-based method that can be used to determine unknown sequences at the 5' and 3' ends of cDNA. Typically, these methods are referred to as 5′ RACE and 3′ RACE, respectively.
In 5' RACE, first-strand cDNAs of any length (even those that do not reach the 5' end of the mRNA) will have additive sequences (i.e., homopolymer tail structures or junctions) that are subsequently amplified by PCR. To maximize the synthesis of full-length cDNAs, a reverse transcription with minimal RNase H activity, high sustained synthesis capacity, and high thermal stability should be selected.
In 3' RACE, the full-length cDNA is not important because no upstream sequence of the PCR start site will be amplified. However, it is preferable to use a reverse transcription that generates long cDNAs, as cDNAs that do not reach the PCR primer binding site will not appear in the RACE analysis.
- gene expression microarray
When selecting a reverse transcription for the preparation of cDNA targets for use in gene chip experiments, the ability to obtain high yields of full-length cDNAs is critical for good coverage of RNAs, including GC-rich or RNA sequences with secondary structures. Equally important, to ensure that a high signal-to-noise ratio is obtained, the reverse transcription must be able to efficiently integrate modified nucleotides, allowing for accurate and unbiased detection of the starting RNA.
- RNA Sequencing (RNA-Seq)
With the advent of second-generation sequencing (NGS), RNA-Seq has become a high-throughput method for analyzing the whole transcriptome (i.e., transcribed coding and long noncoding RNAs), determining gene expression, discovering splice variants and fusion transcripts, and detecting low abundance genes. Advantages of RNA-Seq over gene chips include greater dynamic range, higher sensitivity, and the ability to characterize RNA sequences without genomic information.
Reverse transcription fidelity may play an important role in situations where sequence accuracy is critical, such as RNA sequencing. Reverse transcription fidelity refers to the sequence accuracy during reverse transcription of RNA to DNA. Fidelity is inversely proportional to the reverse transcription error rate. MMLV-based reverse transcription have been reported to have an error rate of one incorrect nucleotide out of 15,000 to 27,000 nucleotides synthesized, whereas AMV reverse transcription exhibits a higher error rate.
Reverse transcription exhibits terminal nucleotidyl transferase (TdT) activity, which, for RNA sequencing (RNA-Seq), induces reverse transcription to specifically incorporate a series of Cs into the 3′ end of the cDNA. this type of TdT activity is triggered during and post-synthesis stages of cDNA synthesis under high concentrations of magnesium and/or manganese ions. In combination with specially designed DNA oligonucleotides having 3' Gs (called template switching oligonucleotides), TdT activity can specifically modify 3' cDNA ends and 5′ RNA ends. Examples of these sequence modifications include the introduction of restriction sites for subsequent cDNA synthesis steps and/or the addition of junctions for downstream RNA sequencing steps.
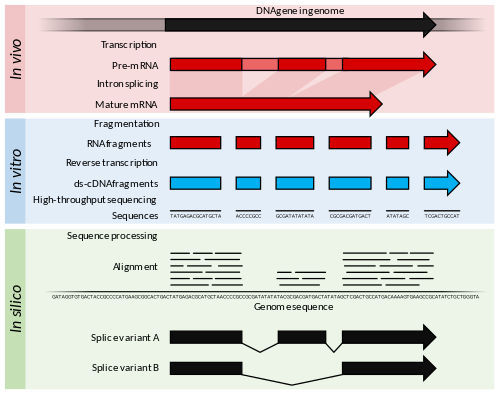
Figure3. Summary of RNA-Seq
Source:https://www.sperikon.com/1235.html


