Ribonuclease inhibitor (RI) is a large (~450 residues, ~49 kDa), acidic (pI ~4.7), leucine-rich repeat protein that forms extremely tight complexes with certain ribonucleases. It is a major cellular protein, accounting for about 0.1% of the weight of all cellular proteins, and appears to play an important role in regulating RNA lifetime.
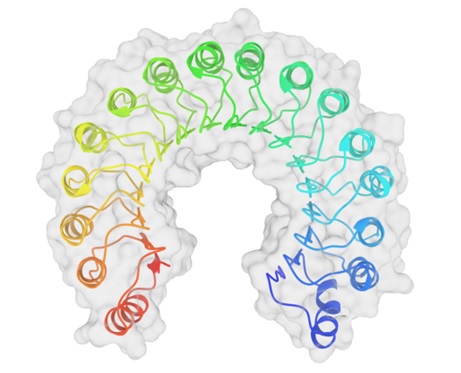
Image 1. Top view of a porcine ribonuclease inhibitor, showing its horseshoe shape. The outer layer is composed of α-helices and the inner layer is composed of parallel β-strands. The inner and outer diameters are approximately 2.1 nm and 6.7 nm, respectively.
RI has a surprisingly high cysteine content (~6.5% vs. 1.7% in typical proteins) and is sensitive to oxidation.RI is also rich in leucine (21.5% leucine vs. 9% in typical proteins) and correspondingly low in other hydrophobic residues (especially leucine). Valine, isoleucine, methionine, tyrosine and phenylalanine.
Structure
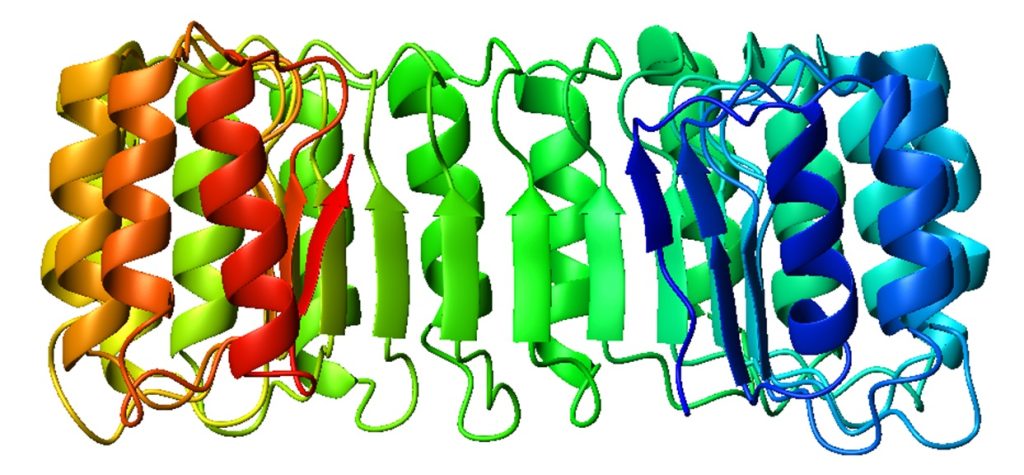
Image 2. Side view of porcine ribonuclease inhibitor; colour band colours from blue (N-terminal) to red (C-terminal).
RIs are classical leucine-rich repeat proteins consisting of alternating alpha helices and beta chains along the main chain. These secondary structure elements surround a curved right-handed solenoid resembling a horseshoe. The parallel β and α helices form the inner and outer walls of the horseshoe, respectively. As it transitions from the α-helix to the β-strand, the structure appears to be stabilised by asparagine buried at the bottom of each corner. αβ repeat lengths alternate between 28 and 29 residues, effectively forming a 57-residue unit corresponding to its genetic structure (each exon encodes a 57-residue unit).
Binds to ribonuclease
The affinity of RI for ribonucleases is the highest of all protein-protein interactions; under physiological conditions, the dissociation constant of the RI-RNase A complex is in the femtomolar (fM) range. Despite its high affinity, RI is able to bind to multiple RNase A species despite their relatively low sequence identity. Both biochemical studies and crystal structures of the RI-RNase A complex indicate that this interaction is primarily controlled by electrostatic interactions but also involves substantial buried surface area. The affinity of RI for ribonucleases is important because many ribonucleases have cytotoxic and cytostatic effects that are closely related to the binding ability of RI.
Mammalian RIs are unable to bind certain pancreatic ribonuclease family members from other species. In particular, amphibian RNases, such as lampinases and amphoteric enzymes from the northern leopard frog, can evade mammalian RI and have been noted to have differential cytotoxicity against cancer cells.
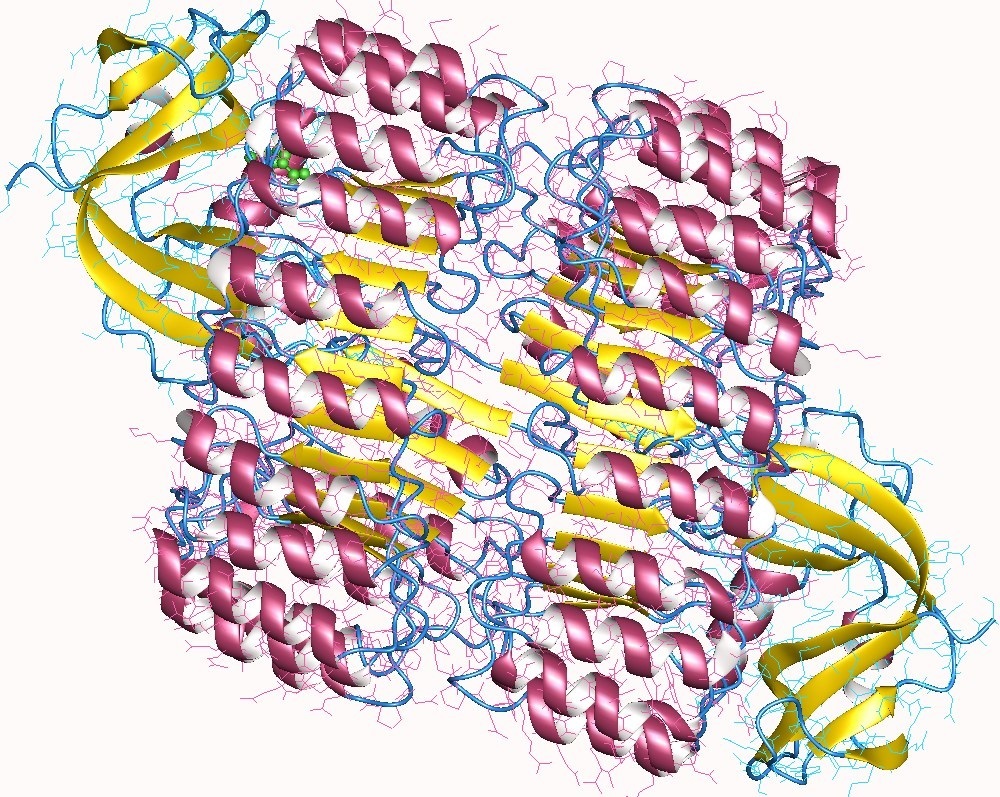
Image 3. RNase I (yellow) and inhibitor (pink helix) complex heterotetramer, human.
Source
https://en.wikipedia.org/wiki/Ribonuclease_inhibitor