Restriction endonucleases (also known as restriction endonucleases) are enzymes that can recognize and attach to specific nucleotide sequences and bind to the phosphodiester between two deoxyribonucleotides at specific locations in each strand. A type of enzyme that cuts bonds is called a restriction enzyme. According to the structure of restriction enzymes, the required cutting sites and modes of action of cofactors, restriction enzymes can be divided into three types: Type I, Type II and Type III. ). Type I restriction endonucleases can catalyze both methylation of host DNA and hydrolysis of unmethylated DNA, while type II restriction enzymes can only catalyze the hydrolysis of unmethylated DNA. Type III restriction endonucleases have both modification and cognitive cleavage functions.
Definition
A class of enzymes that recognize specific deoxyribonucleotide sequences and cleave the phosphodiester bond between two deoxyribonucleotides at specific locations in each chain.
Source
It is generally composed of the first letter of the microbial genus name and the first two letters of the species name, and the fourth letter indicates the strain (strain). For example, the restriction endonuclease extracted from Bacillus amylolique faciens H is called Bam H. Several enzymes with different specificities that recognize different base sequences obtained from the same strain of bacteria can be assigned different numbers, such as HindII. , HindⅢ, HpaI, HpaⅡ, MboI, MboⅡ, etc.
Enzyme reaction: Restriction endonuclease can split the DNA molecule at a limited number of specific sites. It recognizes foreign DNA and degrades it.
Unit definition: The amount of enzyme that digests 1 μg of lambda DNA in 0.05 mL reaction mixture in 1 hour at the specified pH and 37°C is 1 unit.
The product does not contain non-specific nucleic acid hydrolases (indicated by the stability of the gel electrophoresis pattern obtained by incubating 10 units of endonuclease with 1 μg λ DNA for 16 hours). This type of enzyme is mainly isolated from prokaryotes. About 4,000 restriction enzymes have been isolated so far from nearly 300 different microorganisms.
Distribution Area
Restriction endonucleases are widely distributed. At least one restriction endonuclease is found in almost all bacterial genera and species, and many of them have dozens of species in one genus, such as in Haemophilus ( Haemophilus), 22 species have been discovered. Some strains contain very low enzyme content and are difficult to isolate and characterize; however, in some strains, the enzyme content is extremely high. For example, pMB4 of E. coli (EcoRI enzyme) and H. aegyptius (isolate and purify the amount of enzyme that can digest 10g of lambda phage DNA. Bacteria are restriction endonucleases, especially type I restriction enzymes with very strong specificity. primary sources.
Classification Properties
Restriction enzymes can be divided into two categories based on the functional characteristics, size and cofactors required for the reaction, namely Class I enzymes and II enzymes. EcoK and EcoB, which were first discovered in E. coli, belong to class I enzymes. Its molecular weight is relatively large; in addition to Mg2+, S-adenosyl-L methionine and ATP are also required during the reaction; there are no specific enzymatic fragments on the DNA molecule, which is the most common among Class I and Class II enzymes. Significant differences. Therefore, class I enzymes are of little value as DNA analysis tools. Class II enzymes include EcoR I, BamH I, Hind II, Hind III, etc. Its molecular weight is less than 105 Daltons; the reaction only requires Mg2+; the most important thing is that it has a specific cutting point on the specific base sequence recognized, so the DNA molecule can undergo specific enzymatic hydrolysis after being acted upon by Class II enzymes. Fragments can be separated and identified by gel electrophoresis.
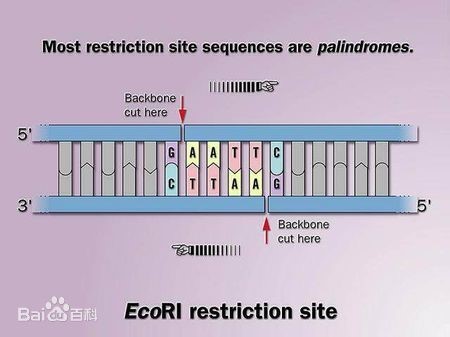
Restriction enzymes recognize palindromic sequences in DNA sequences. The cleavage site of some enzymes is on one side of the palindrome (such as EcoR I, BamH I, Hind, etc.), thus forming sticky ends. Other class II enzymes such as Alu I, BsuR I, Bal I, Hal III, HPa I, Sma I, etc., the cutting site is in the middle of the palindromic sequence, forming a blunt end. The cleavage site of Alu I is as follows:
5'-A G^C T-3'
3'-T C^G A-5'
Among the discovered restriction enzymes, the recognition sequences of nearly one hundred enzymes have been determined. There are many enzymes from different sources that have the same base recognition sequence. Such enzymes are called "isozoomers" (isochizomers, iso-restriction enzymes; iso-cleavage enzymes). It should be noted that although these enzymes have the same recognition sequence, their cutting points are not exactly the same. For example, both Xma I and Sma I recognize the hexanucleotide CCGGGG, but the cutting point of Xma I is at CCCCGGG, while the cutting point of Ema I is at CCCGGGG. The former cuts DNA molecules to form DNA fragments with CCGG sticky ends, while the latter Does not form sticky ends (called flat ends). Of course, there are also enzymes with the same recognition sequence and cleavage point, such as Hap Ⅱ, Hpa Ⅱ, and Mno I, which all have the same cleavage point in the recognition sequence CCGG. Hal III and BsuR I also have the same cleavage point in the recognition sequence GGCC. point.
Application
Used for the construction of physical maps of DNA genomes; gene positioning and gene isolation; DNA molecule base sequence analysis; comparison of related DNA molecules and genetic engineering; and genetic engineering editing.
Restriction endonucleases are produced by bacteria, and their physiological significance is to improve their own defense capabilities.
Restriction enzymes generally do not cut their own DNA molecules, but only cut foreign DNA.
Name
The naming of restriction enzymes is based on the bacterial species, taking EcoRI as an example:
Naming of restriction enzymes
| E | Escherichia | (genus) |
| co | coli | (type) |
| R | RY13 | (strain) |
| I | First to discover | The order found in this class of bacteria |
Type
type I restriction enzyme
It has the functions of modification and restriction at the same time; it also has the ability to recognize specific base sequences on DNA. Usually the cleavage site is thousands of miles away from the recognition site. base away.
For example: EcoB, EcoK.
type II restriction enzyme
It only has the function of recognition and cleavage, and the modification is performed by other enzymes. Most of the recognized positions are short palindrome sequences; the cut base sequence is usually the recognized sequence. It is a highly practical type of restriction enzyme in genetic engineering.
For example: EcoRI, HindIII.
Type III restriction enzyme
Similar to the first type of restriction enzyme, it has the functions of modification and recognition and cleavage at the same time. It can recognize short asymmetric sequences, and the distance between the cutting site and the recognition sequence is about 24-26 base pairs.
For example: HinfⅢ.
Physiological Significance
Restriction is actually a protective mechanism that restricts enzymes to degrade foreign DNA and maintain host genetic stability. Methylation is a common modification that protects adenine A and cytosine C. Methylation is used to identify one's own genetic material and foreign genetic material. Therefore, bacteria that can produce restriction enzymes that defend against virus infection may have sequences recognized by the enzyme in their own genomes, but the recognition sequences or enzyme cutting sites are methylated. But this does not mean that once it is methylated, all restriction enzymes cannot cut it. Most restriction enzymes are sensitive to DNA methylation, so when the target sequence of the restriction enzyme overlaps with the methylation site, there are three possibilities for the impact on the enzyme digestion, namely no impact, partial impact, and complete blockage. The ability to cleave methylated DNA is an intrinsic and unpredictable property of restriction enzymes; therefore, to efficiently cleave DNA, both DNA methylation and the sensitivity of the restriction enzyme to that type of methylation must be considered. Additionally, most commercial restriction enzymes today are specifically designed to cleave methylated DNA.
Source: https://baike.baidu.com/


